Download the vHelix plugin for Autodesk Maya
The plugin is available for the 64-bit versions of Autodesk Maya 2011, 2012, 2013, 2014 , 2016, 2018 and 2019 under Windows and Mac OS X. It is currently not available for other versions of Maya and not for the 32-bit versions. The plugin need to be compiled in order to work with other versions of Maya. If you are interested in using the plugin with another version of Autodesk Maya, you will need to download the source from https://github.com/gardell/vHelix and compile it for your system.
 Windows - Please note the install instructions for Maya 2016-2019
Windows - Please note the install instructions for Maya 2016-2019
The windows build (< Maya 2019) are compiled using Microsoft Visual Studio 2013, the version for Maya 2019 is compiled using Microscoft Visual Studio 2019. This requires that you install the Microsoft Visual C++ Redistributable Package (x64) available at http://www.microsoft.com/en-us/download/details.aspx?id=40784 ( Maya < 2019) or https://support.microsoft.com/en-us/help/2977003/the-latest-supported-visual-c-downloads (Maya 2019) or as an optional software update on Windows/Microsoft update. A debug version of the software (requires Microsoft Visual Studio 2013 to be installed) is available on the github page. Release versions:
Download: Maya 2012 Windows x64 | Maya 2013 Windows x64 | Maya 2014 Windows x64 | Maya 2016 Windows x64 | Maya 2018 Windows x64 | Maya 2019 Windows x64 | Install Instructions
 Linux
Linux
The linux build was compiled under Ubuntu 13.10 64-bit using gcc 4.8.1
Maya 2014 Linux x64 | Documentation
 Mac OS X - Please note the install instructions for maya 2016-2019
Mac OS X - Please note the install instructions for maya 2016-2019
The Mac OS X build was compiled using gcc 4.2 targeted at 64-bit Mac OS X 10.8 and newer.
Maya 2013 Mac OS X x64 | Maya 2014 Mac OS X x64 | Maya 2016 Mac OS X x64 | Maya 2018 Mac OS X x64 | Maya 2019 Mac OS X x64 | Install Instructions
vHelix comes from the lab of Björn Högberg at Karolinska Institutet. Design by Björn Högberg and Johan Gardell, programmed by Johan Gardell.
Automated DNA nanostructure design from polyhedral meshes
vHelix is developed to be used in conjunction with the method reported in the paper [Benson et. al “DNA Rendering of Polyhedral Meshes at the Nanoscale” Nature (2015) DOI 10.1038/nature14586].
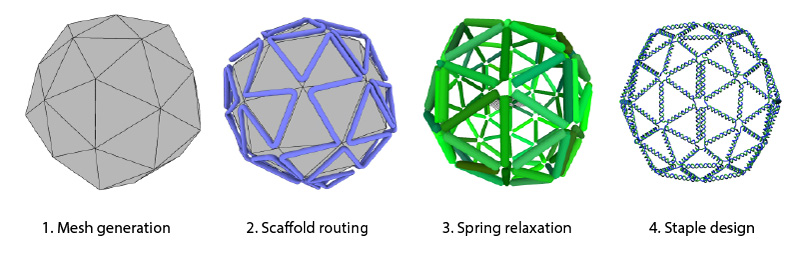
The process starts with the design of a polyhedral mesh in a 3D computer graphics software and exporting it in the PLY format. The mesh is then run through the BSCOR software to generate a scaffold path and relaxed physical DNA representation of the mesh. BSCOR can be downloaded and run on your local machine (recommended for performance). Alternately, we offer an online version of BSCOR where the mesh processing is performed on our server . This process produces a file in the .rpoly format that can be imported to vHelix for staple design.
The BSCOR software package for converting polyhedral meshes into DNA nanostructure designs can be downloaded here. The package consists of a scaffold routing algorithm and a spring relxation software. This package also supports the routing of open 2D meshes as described in DOI: 10.1002/anie.201602446
The scaffold routing algoritm was developed by the Orponen group at Aalto University, the source code is available on https://github.com/mohamma1/bscor
The spring relaxation was developed by the Högberg lab and the source code is available from https://github.com/gardell/scaffold-routing-rectification
Please wach our tutorials below for an introduction on how to use the design paradigm on your own.
If you have any trouble running the software, please contact erik.benson [at] physics.ox.ac.uk
Simulation of vHelix designs
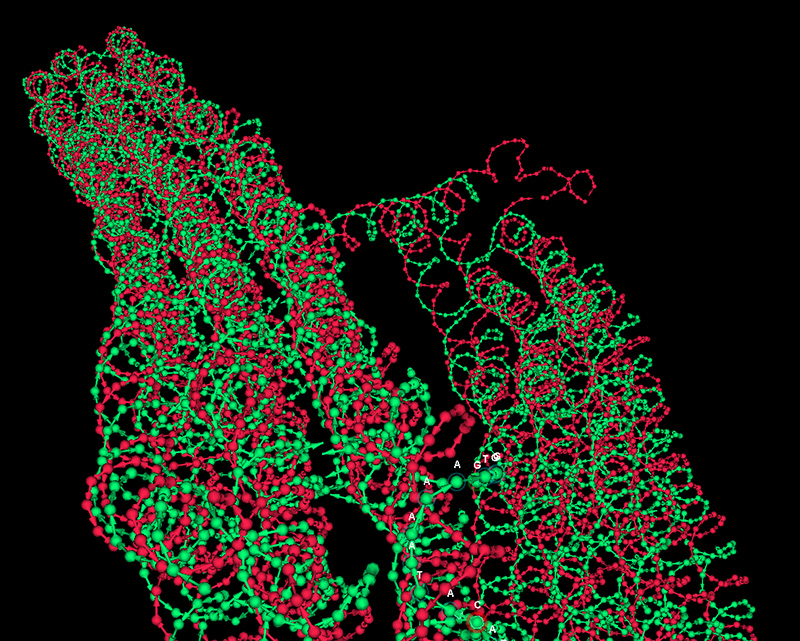
oxDNA
oxDNA is a powerful coarse-grained molecular dynamics simulation system for DNA and has been widley used to simulate DNA nanostructures. vHelix is a usefull starting point for the simulation of both small systems and large DNA origami structures (either from wireframes or cadnano lattice designs).
DNA designs in vhelix saved in a maya file in the Maya Ascii format (.MA) can be converted to oxdna start files using the tacoxDNA server http://tacoxdna.sissa.it/vHelix_oxDNA.
Alternativley, the maya 2019 version of vHelix has a built in oxDNA exporter that is accesible trough the maya menu File > Export All . This exporter will generate an extra meta-datafile called .vhelix that can be used to reimport a DNA system after simulation in oxDNA (experimental).
More documentation to follow...
mrDNA
mrDNA is a recently introduced software package for multi-resoultion simulation of DNA nanostructures https://www.biorxiv.org/content/early/2019/12/05/865733.full.pdf . mrDNA supports simulating vhelix designs, including pulling them throud nano-capillaries.
Tutorials
vHelix tutorial 1
DNA nanostructures from polyhedra meshes 1
vHelix tutorial 2
DNA nanostructures from polyhedral meshes 2
Vhelix for oxDNA tutorials
Tutorial 1: Getting started with oxDNA on windows
Tutorial 2: creating small DNA systems in vhelix and simulating in oxDNA
Tutorial 3: Creating a larger rotaxane system in vHelix for simulation in oxDNA
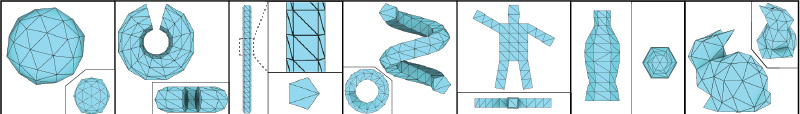
Example mesh bases structures
Citing vHelix
Please use this reference when citing vHelix:
Benson et. al “DNA Rendering of Polyhedral Meshes at the Nanoscale” Nature (2015) DOI 10.1038/nature14586
